1. Manifest
The Process Macro by Andrew F. Hayes has helped thousands of researchers in their analysis of moderation, mediation, and conditional processes. Unfortunately, Process was only available for proprietary softwares (SAS and SPSS), which means that students and researchers had to purchase a license of those softwares to make use of the Macro.
Because of the growing popularity of Python in the scientific community, I decided to implement the features of the Process Macro into an open-source library, that researchers will be able to use without relying on proprietary softwares. pyprocessmacro is open-source, and released under a MIT license.
2. Features
A. Current Implementation
In the current version, pyprocessmacro replicates the following features from the original Process Macro v2.16:
- All models (1 to 76), with the exception of Model 6 (serial mediation)
are supported, and have been numerically tested for accuracy against the
output of the original Process macro (see the
test_models_accuracy.py) - Estimation of binary/continuous outcome variables. The binary outcomes are estimated in Logit using the Newton-Raphson convergence algorithm, the continuous variables are estimated using OLS.
- Estimation of (conditional) direct and indirect effects
- Indices for Partial/Conditional/Moderated Moderated Mediation
- Automatic generation of spotlight values for continuous/discrete moderators.
- Rich set of options to tweak the estimation and display of the different models: (almost) all the options from Process exist in pyprocessmacro. See the documentation for more details.
B. Changes and Improvements
The following changes and improvements have been made from the original Process Macro:
- Variable names can be of any length, and even include spaces and special characters.
- All mediation models support an infinite number of mediators (versus a maximum of 10 in Process).
- Normal theory tests for the indirect effect(s) are not reported, as the bias-corrected bootstrapping approach is now widely accepted and in most cases more robust.
- Plotting capabilities: pyprocessmacro can generate the plot of direct and indirect effects at various levels of the moderators.
- Fast estimation process: pyprocessmacro leverages the capabilities of NumPy to efficiently compute a large number of bootstrap estimates, which dramatically speeds up the estimation of complex models.
- Transparent bootstrapping: pyprocessmacro automatically store the bootstrap estimates for convenient inspection, and explicitely reports the samples that have been discarded because of numerical instability.
C. Missing Features
In the current version, the following features have not yet been ported to pyprocessmacro:
- Support for categorical independent variables.
- Generation of individual fixed effects for repeated measures.
- R² improvement from moderators in moderation models (1, 2, 3).
- Estimation of serial mediation (Model 6)
- Some options (
normal,varorder, ...). pyprocessmacro will issue a warning if you are trying to use an option that has not been implemented.
3. Installation and Usage
A. Installation
You can install pyprocessmacro with pip:
pip install pyprocessmacro
B. Minimal example
A Process object is initialized by specifying a data source, the
model number, and the mapping between the variable names in the data
and the symbols used by the model.
from pyprocessmacro import Process
import pandas as pd
df = pd.read_csv("MyDataset.csv")
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"])
p.summary()
You might have noticed that there is no argument varlist in
pyprocessmacro: this list is automatically inferred from the variable
names given to x, y, m. Other than this, the syntax for pyprocessmacro
is (almost) identical to that of Process. Unless this documentation
mentions otherwise, you can assume that all the options/keywords from
Process exist in pyprocessmacro.
Once the object is initialized, its summary() method is used to
display the estimation results.
C. Adding Statistical Controls
In Process, the controls are defined as "any argument in the varlist that is not the IV, the DV, a moderator, or a mediator." In pyprocessmacro, the list of variables to include as controls have to be explicitely specified in the "controls" argument.
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"],
controls=["Control1", "Control2"],
controls_in="all")
p.summary()
The equation(s) to which the controls are added is specified through
a controls_in argument:
x_to_mmeans that the controls will only be added in the path from the IV to the mediator(s) only.all_to_ymeans that the controls will only be added in the path from the IV and the mediators to the DV.allmeans that the controls will be added in all equations.
The ability to specify a different list of control for each equation is coming in the next release of pyprocessmacro.
D. Logistic Regressions for Binary Outcomes
The original Process Macro automatically uses a Logistic (instead of OLS) regression when it detects a binary outcome.
pyprocessmacro favors a more explicit approach, and requires you to set
the parameter logit to True if your DV should be estimated using a
Logistic regression.
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"], logit=True)
p.summary()
It goes without saying that this will return an error if your DV is not dichotomous.
E. Spotlight Values for Moderators
In Process as in pyprocessmacro, the spotlight values of the moderators are defined as follow:
- By default, the spotlight values are equal to M - 1SD, M and M + 1SD, where M and SD are the mean and standard deviation of that variable.
- If the option
quantile=Trueis specified, then the spotlight values for each moderator are the 10th, 25th, 50th, 75th and 90th percentile of that variable. - If a moderator is a discrete variable, the spotlight values are those discrete values.
In Process, custom spotlight values can be applied to each moderator q, v, z, ... through the arguments qmodval, vmodval, zmodval...
In pyprocessmacro, the user must instead supply custom values for each
moderator in a dictionary passed to the modval parameter:
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"],
modval={
"Motivation":[-5, 0, 5], # Moderator 'Motivation' at values -5, 0 and 5
"SkillRelevance":[-1, 1] # Moderator 'SkillRelevance' at values -1 and 1
})
p.summary()
F. Suppress Output
When the Process object is initialized by Python, it displays various
information about the model (model number, list of variables,
sample size, number of bootstrap samples, etc...). If you wish not to
display this information, just add the argument suppr_init=True when
initializing the model.
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"], suppr_init=True)
p.summary()
4. Accessing the Results
After the Process object is initialized, the following methods
allow you to conveniently recover the different estimates of interest:
A. summary()
This method replicates the output that you would see in Process, and displays the following information:
- Model summaries and parameters estimates for all outcomes (i.e. the independent variable, and the mediator(s)).
- If the model has a moderation, conditional effects at the spotlight values of the moderator(s).
- If the model has a mediation, direct and indirect effects.
- If the model has a moderation and a mediation, conditional direct and indirect effects at values of the moderator(s).
- If those statistics are relevant, indices for partial, conditional, and moderated moderated mediation will be reported.
B. outcome_models
This command gives you individual access to each of the outcome models through a dictionary. This allows you to recover the model and parameters estimates for each outcome.
Each OutcomeModel object has the following methods:
summary()prints the full summary of the model (as Process does).model_summary()returns a DataFrame of goodness-of-fit statistics for the model.coeff_summary()returns a DataFrame of estimate, standard error, corresponding z/t, p-value, and confidence interval for each of the parameters in the model.estimation_resultsgives you access to a dictionary containing all the statistical information of the model.
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"], suppr_init=True)
# The model for the outcome "MediationSkills"
model_medskills = p.outcome_models["MediationSkills"]
# Print the summary for this model
model_medskills.summary()
# Store the DataFrame of estimates into a new variable.
df_params_med1 = model_medskills.coeff_summary()
# Store the R² of the model into a new variable.
med1_R2 = model_medskills.estimation_results["R2"]
Note that the methods are called from the model_medskills object!
If you call p.coeff_summary(), you will get an error.
C. direct_model
When the Process model includes a mediation, the direct effect model can be accessed. It implements the following methods:
summary()prints the full summary of the direct effects, as printed when calling Process.summary().coeff_summary()returns a DataFrame of estimate, standard error, t-value, p-value, and confidence interval for each of the (conditional) direct effect(s).
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"], suppr_init=True)
# The model for the direct effect
direct_model = p.direct_model
# Store the DataFrame of estimates into a variable.
df_params_direct = direct_model.coeff_summary()
Note that the methods are called from the direct_model object!
If you call p.coeff_summary(), you will get an error.
D. indirect_model
When the Process model includes a parallel mediation, the indirect effect model can be accessed as well. It implements the following methods:
summary()prints the full summary of the indirect effects, and other related indices, as printed by Process.summary().coeff_summary()returns a DataFrame of indirect effect(s) and their SE/CI for each of the mediation pathsPMM_index_summary()returns a DataFrame of indices for Partial Moderated Mediation, and their SE/CI, for each of the moderators and mediation paths. If the model does not compute a PMM, this will return an error.CMM_index_summary()returns a DataFrame of indices for Conditional Moderated Mediation, and their SE/CI, for each of the moderators and mediation paths. If the model does not compute a CMM, this will return an error.CMM_index_summary()returns a DataFrame of indices for Moderated Moderated Mediation, and their SE/CI, for each of the mediation paths. If the model does not compute a MMM, this will return an error.
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"], suppr_init=True)
# The model for the direct effect
indirect_model = p.indirect_model
# Store the DataFrame of estimates into a variable.
df_params_direct = indirect_model.coeff_summary()
Note that the methods are called from the indirect_model object!
If you call p.coeff_summary(), you will get an error.
5. Recover Bootstrap Estimates
The original Process macro allows you to save the parameter estimates
for each bootstrap sample by specifying the save keyword. The Macro
then returns a new dataset of bootstrap estimates.
In pyprocessmacro, this is done by calling the method
get_bootstrap_estimates(), which returns a DataFrame containing
the parameters estimates. Each row represents a bootstrap repetition,
and the columns are explicitely labeled to identity the variables and
the model.
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"], suppr_init=True)
boot_estimates = p.get_bootstrap_estimates() # Called from the Process object directly.
6. Plotting Capabilities
pyprocessmacro allows you to plot the conditional direct and indirect effect(s), at different values of the moderators.
The methods plot_indirect_effects() and plot_direct_effects() are
identical in syntax, with one small exception: you must specify the name
of the mediator for plot_indirect_effects as a first argument.
They return a seaborn.FacetGrid object that can be used to further
tweak the appearance of the plot.
A. Basic Usage
When plotting conditional direct (and indirect) effects, the effect is always represented on the y-axis. The various spotlight values of the moderator(s) can be represented on several dimensions:
- On the x-axis (moderator passed to
x). - As a color-code, in which case several lines are displayed on the same
plot (moderator passed to
hue). - On different plots, displayed side-by-side (moderator passed to
col). - On different plots, displayed one below the other (moderator passed
to
row)
At the minimum, the x argument is required, while the hue, col
and row are optional.
The examples below are showing what the plots could look like for a
model with two moderators.
from pyprocessmacro import Process
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv("MyDataset.csv")
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation",
z="SkillRelevance", m=["MediationSkills", "ModerationSkills"],
suppr_init=True)
# Conditional direct effects of Effort, at values of Motivation (x-axis)
g = p.plot_direct_effects(x="Motivation")
plt.show()
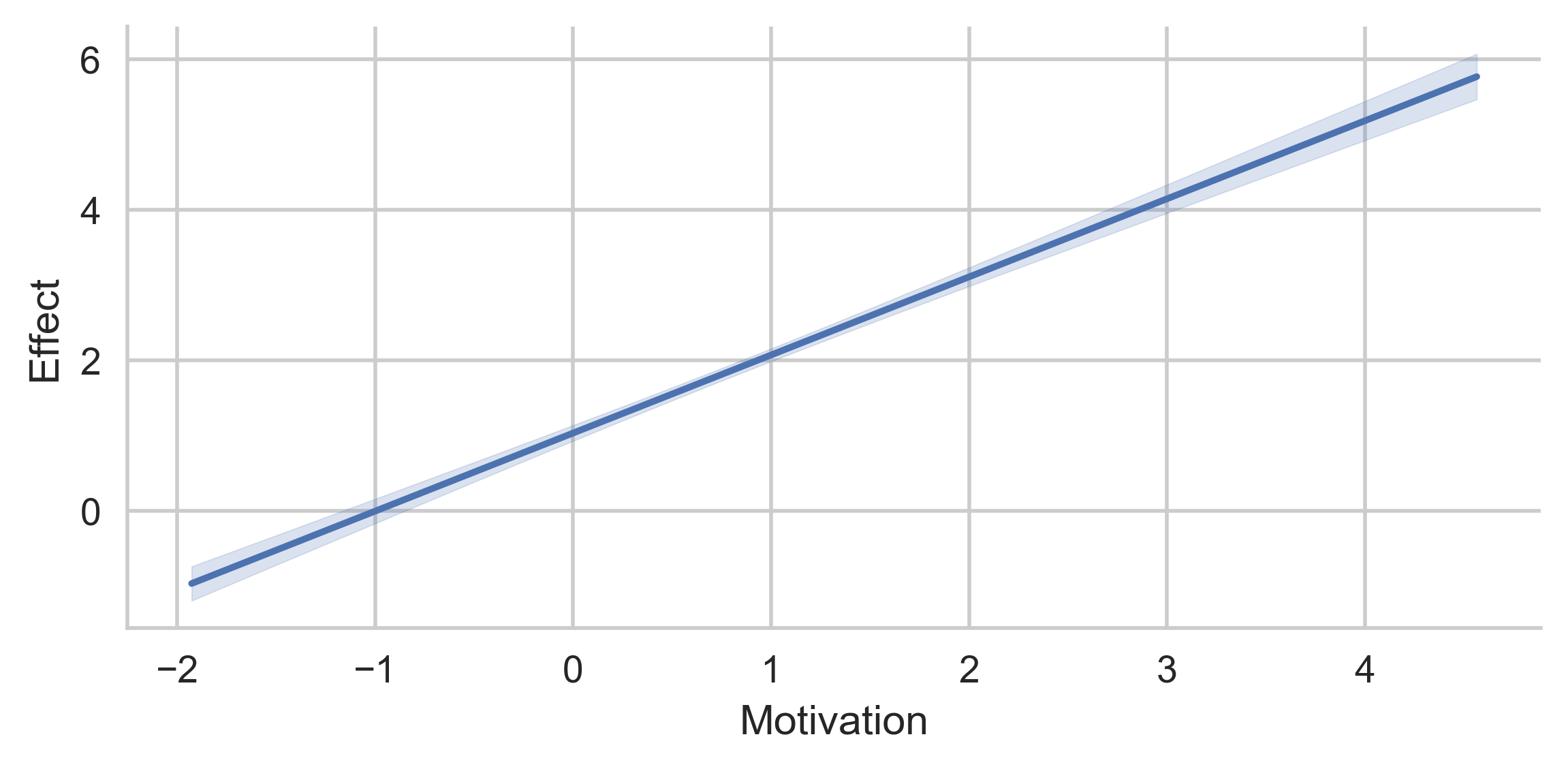
# Conditional indirect effects through MediationSkills, at values of Motivation (x-axis) and
# SkillRelevance (color-coded)
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation", hue="SkillRelevance")
g.add_legend(title="") # Add the legend for the color-coding
plt.show()
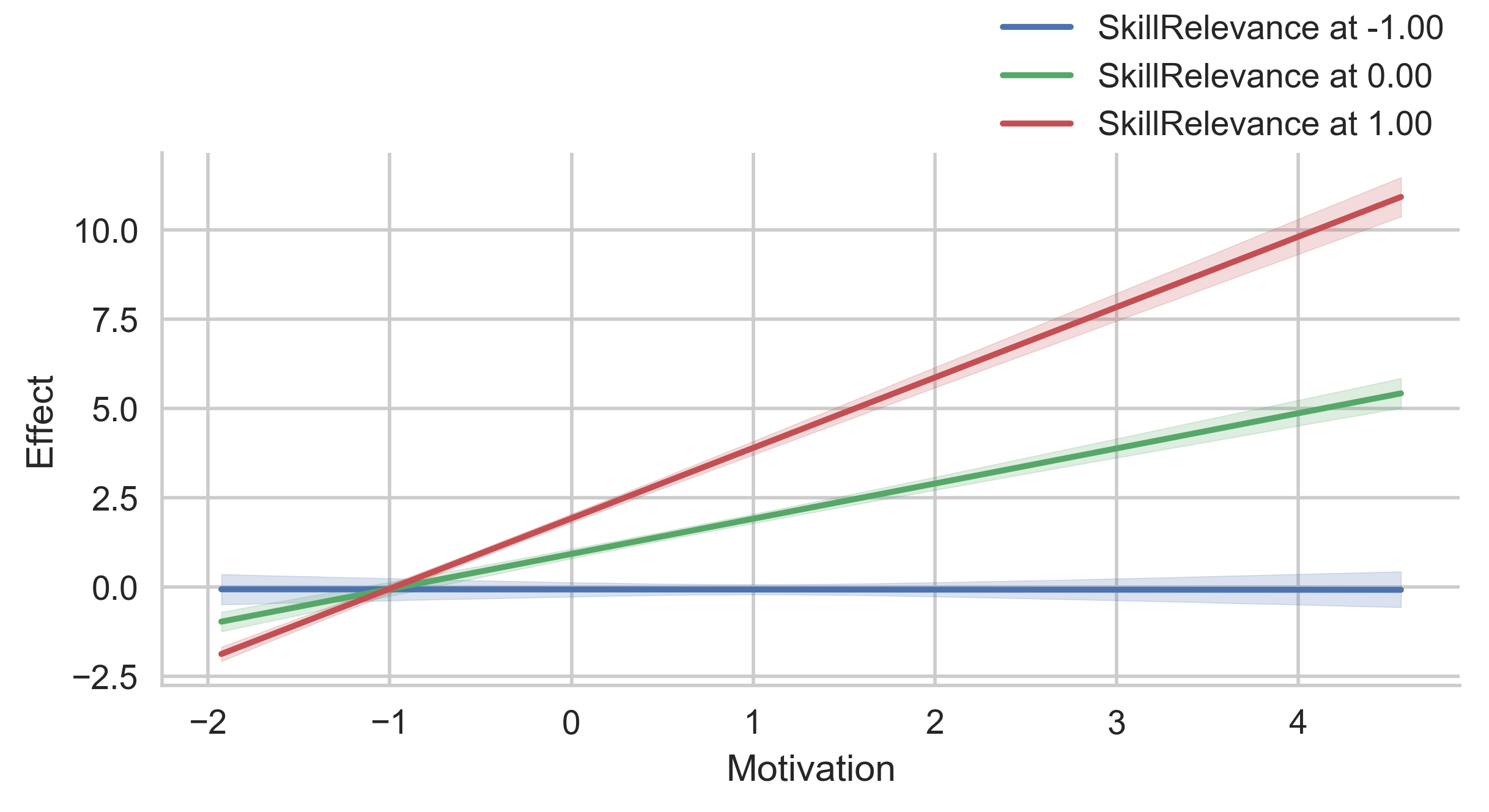
# Display the values for SkillRelevance on side-by-side plots instead.
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation", col="SkillRelevance")
plt.show()
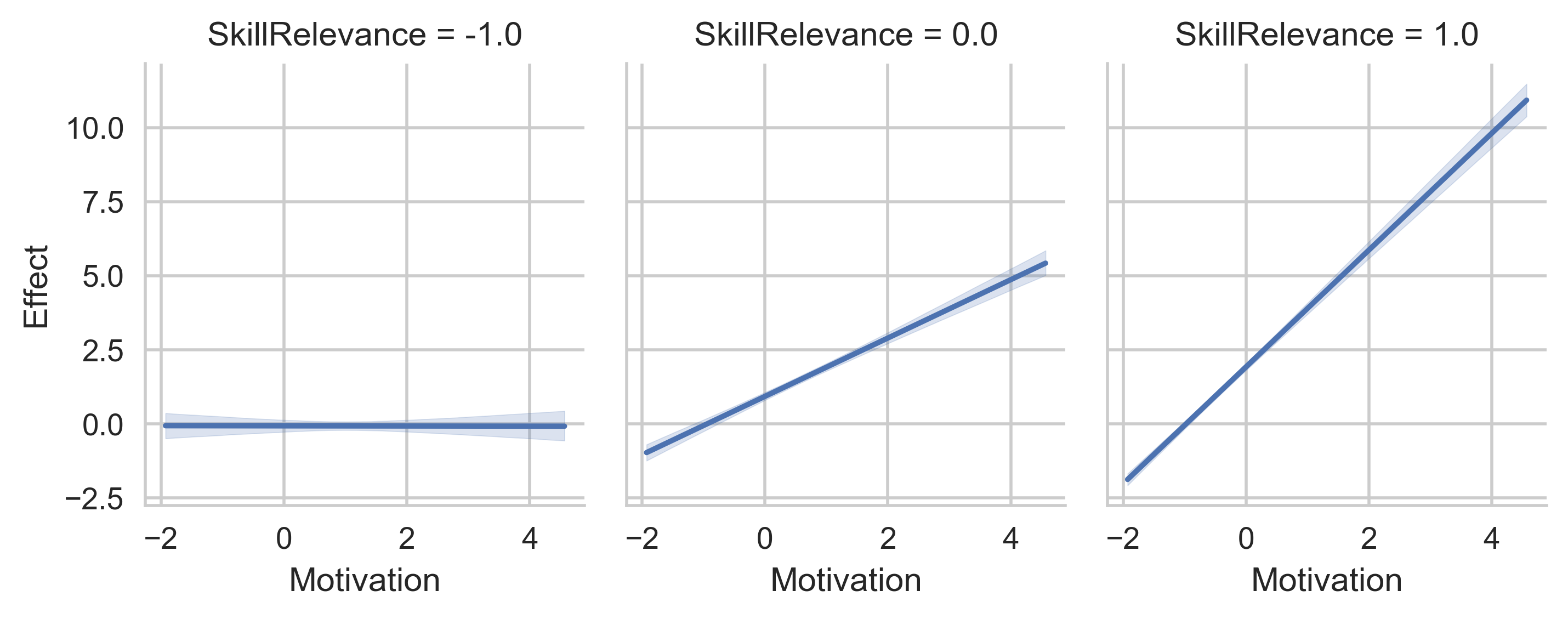
# Display the values for SkillRelevance on vertical plots instead.
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation", row="SkillRelevance")
plt.show()
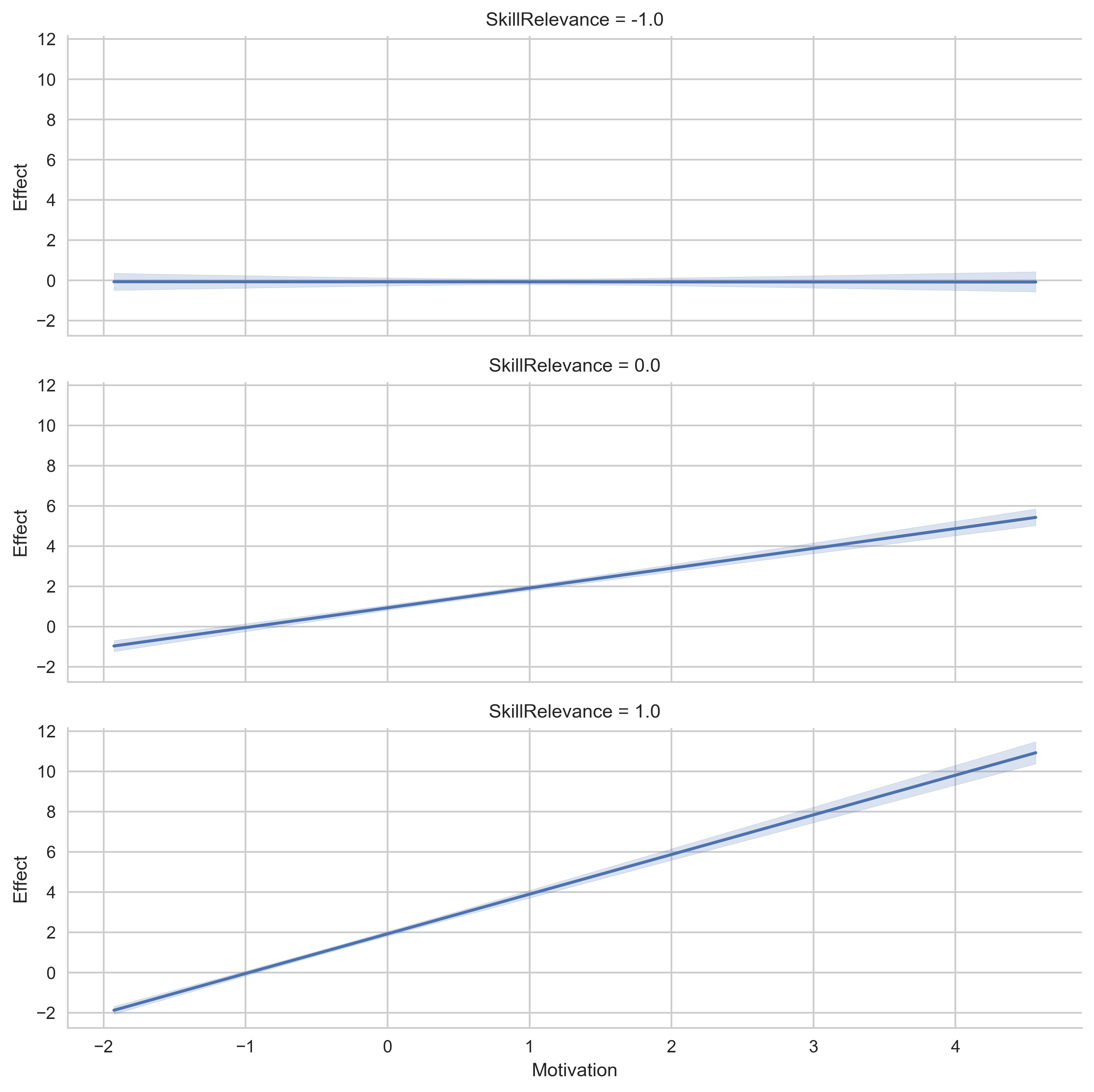
B. Change Spotlight Values
By default, the spotlight values used to plot the effects are the same
as the ones passed when initializing Process.
However, you can pass custom values for some, or all, the moderators
through the mods_at argument. The library will automatically compute
the conditional effects at those new spotlight values, and plot them.
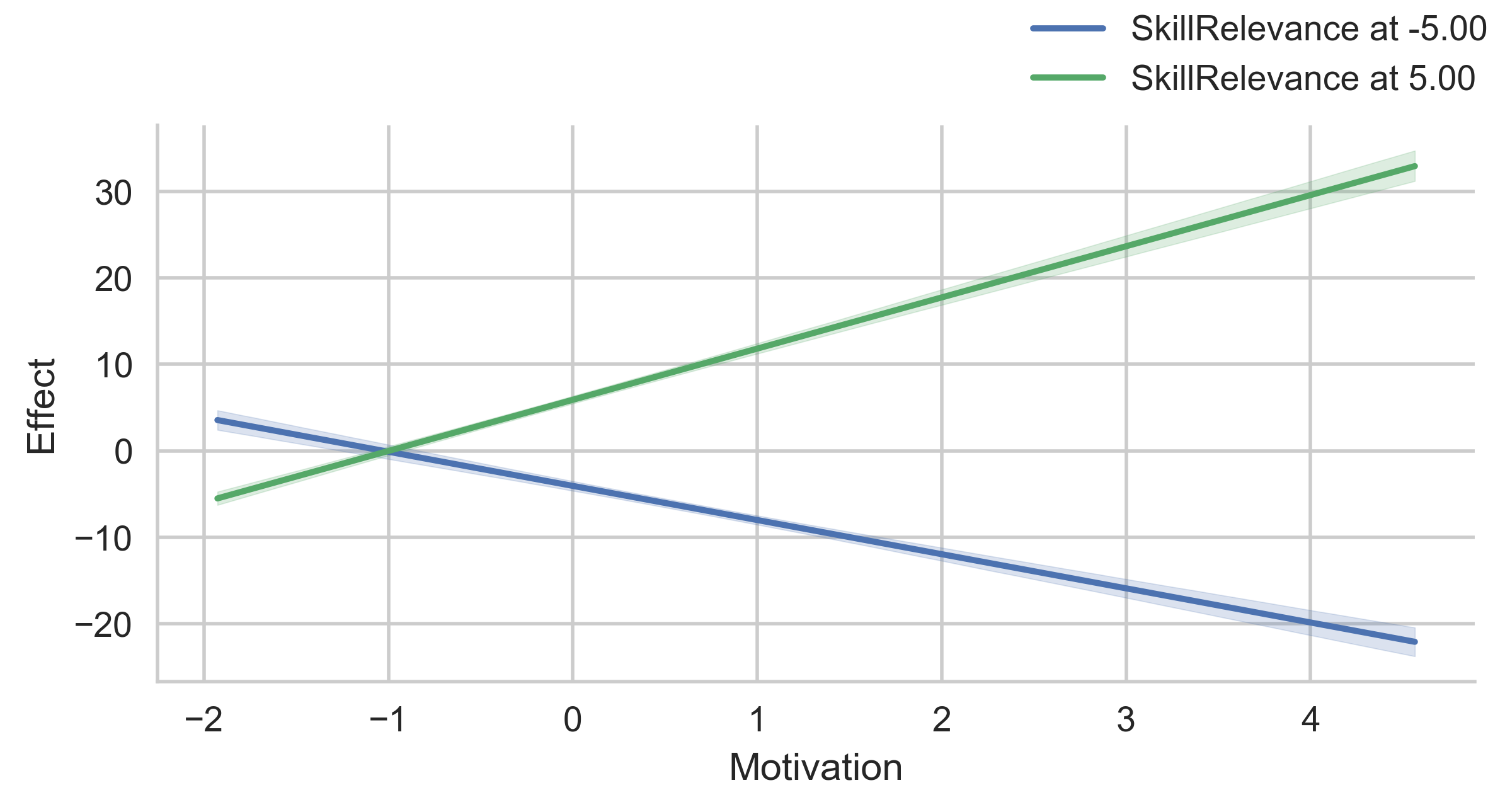
# Change the spotlight values for SkillRelevance
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation", hue="SkillRelevance",
mods_at={"SkillRelevance": [-5, 5]})
g.add_legend(title="")
plt.show()
C. Representation of uncertainty
The display of confidence intervals for the direct/indirect effects can
be customized through the errstyle argument:
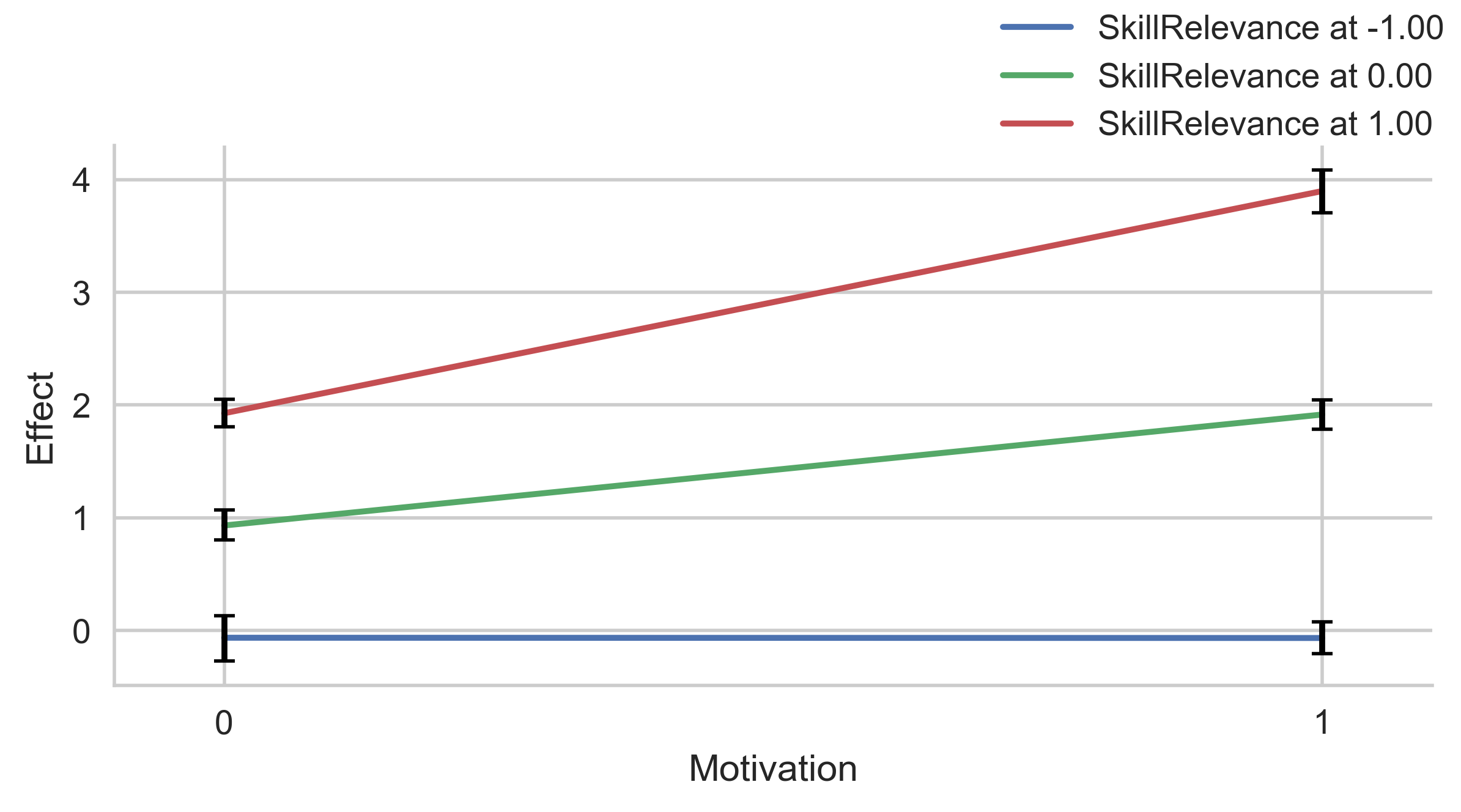
errstyle="band"(default) plots a continuous error band between the lower and higher confidence interval. This representation works well when the moderator displayed on the x-axis is continuous (e.g. age), as it allows you to visualize the error at all levels of the moderator.errstyle="ci"plots an error bar at each value of the moderator on x-axis. It works well when the moderator displayed on the x-axis is dichotomous or has few values (e.g. gender), as it reduces clutter.errstyle="none"does not show the error on the plot.
# CI for dichotomous moderator
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation", hue="SkillRelevance",
mods_at={"Motivation": [0, 1], "SkillRelevance":[-1, 0, 1]},
errstyle="ci")
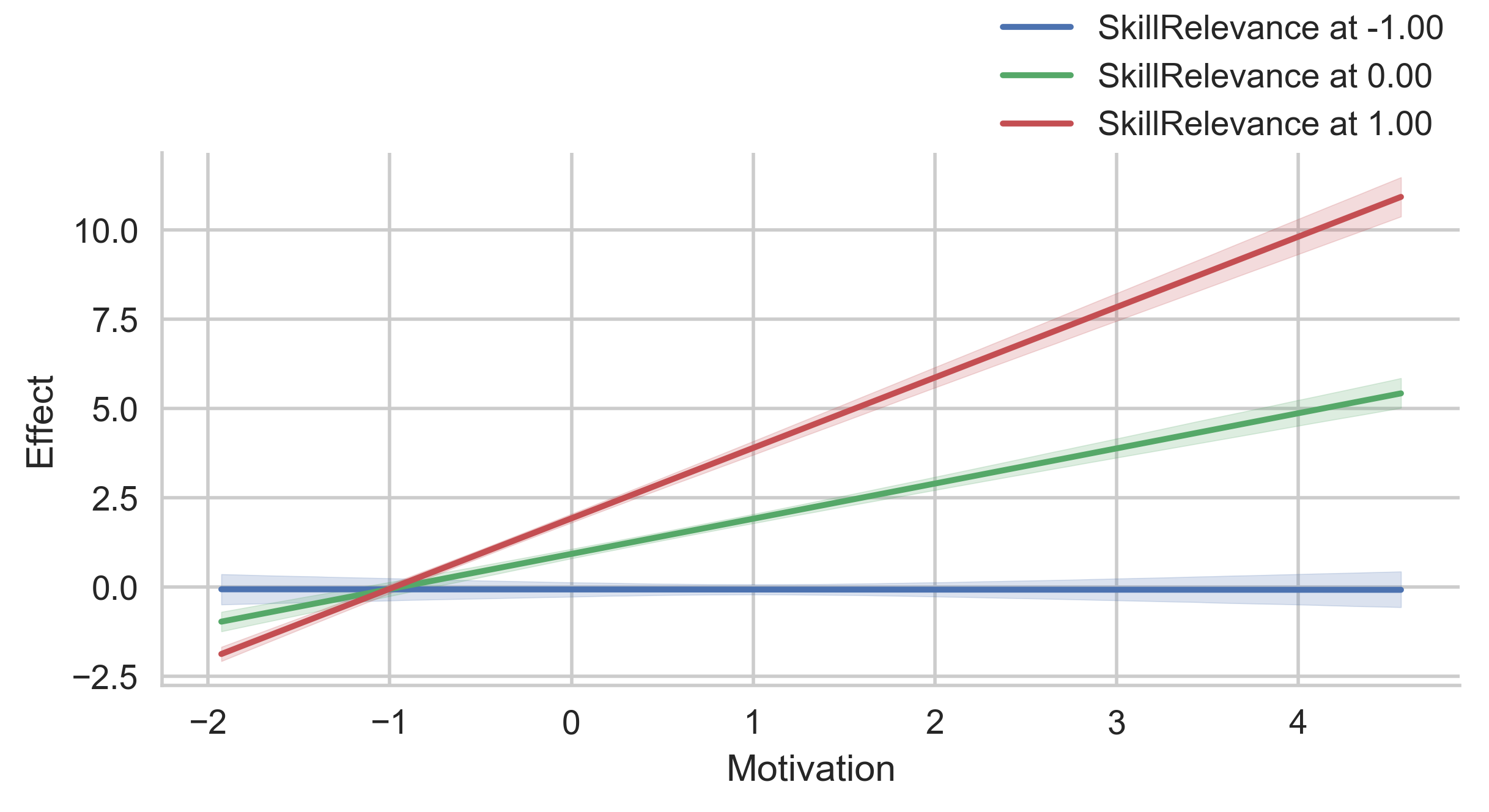
# Error band for continous moderator
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation", hue="SkillRelevance",
mods_at={"SkillRelevance":[-1, 0, 1]},
errstyle="ci")
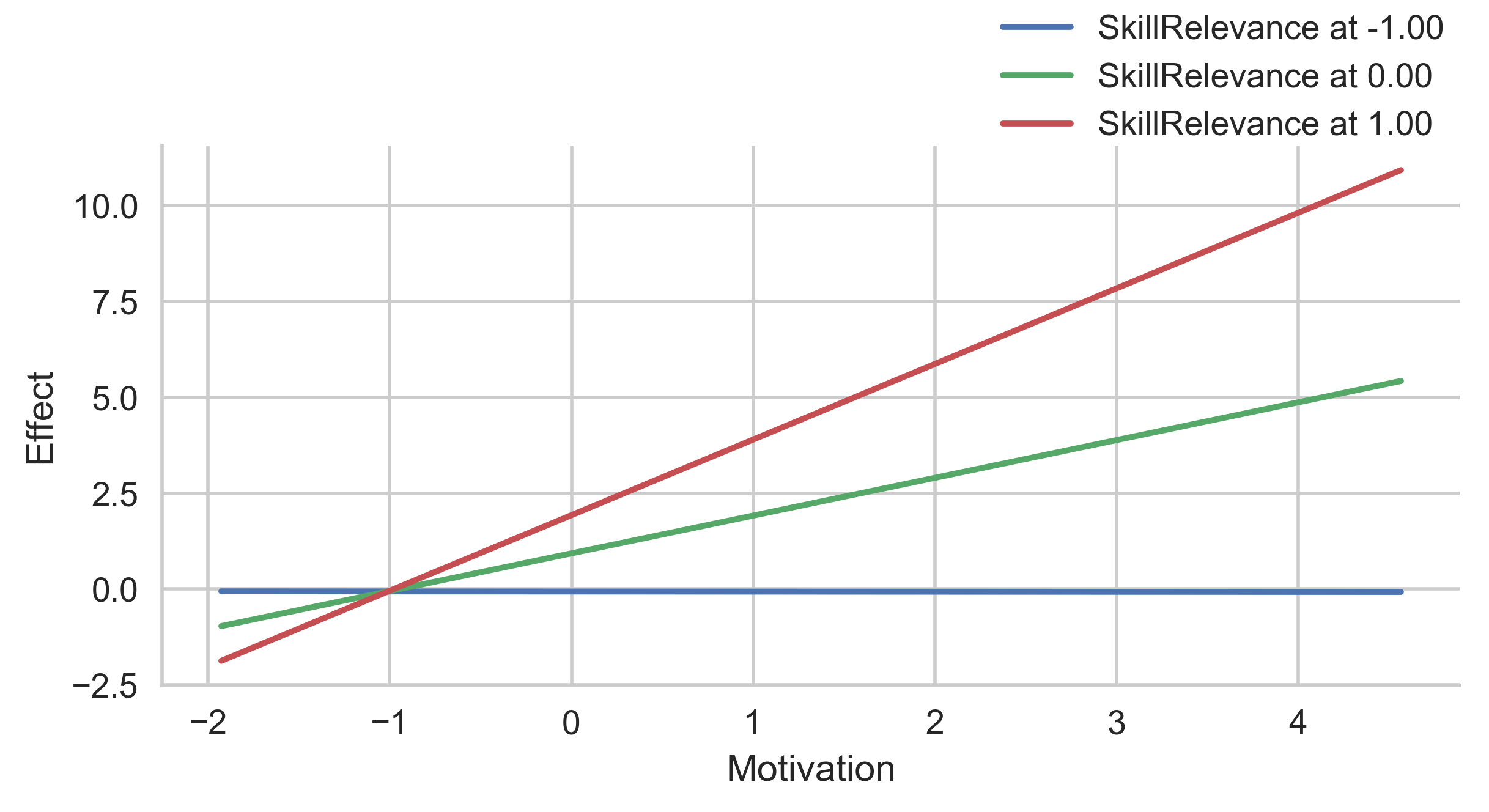
# No representation of error
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation", hue="SkillRelevance",
mods_at={"SkillRelevance":[-1, 0, 1]},
errstyle="none")
plt.show()
D. "Partial" plots
In the example above, the number of moderators supplied as arguments was always equal to the number of moderators in the direct/indirect path.
You can also "omit" some moderators, and plot "partial" conditional direct/indirect effects. In that case, the omitted moderators will assume a value of 0 when computing the direct/indirect effects.
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"], suppr_init=True)
# SkillRelevance is a moderator of the indirect path, but is not mentioned as an argument
# in the plotting function!
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation")
plt.show()
This plot represents the "partial" conditional indirect effect, when SkillRelevance is evaluated at 0:
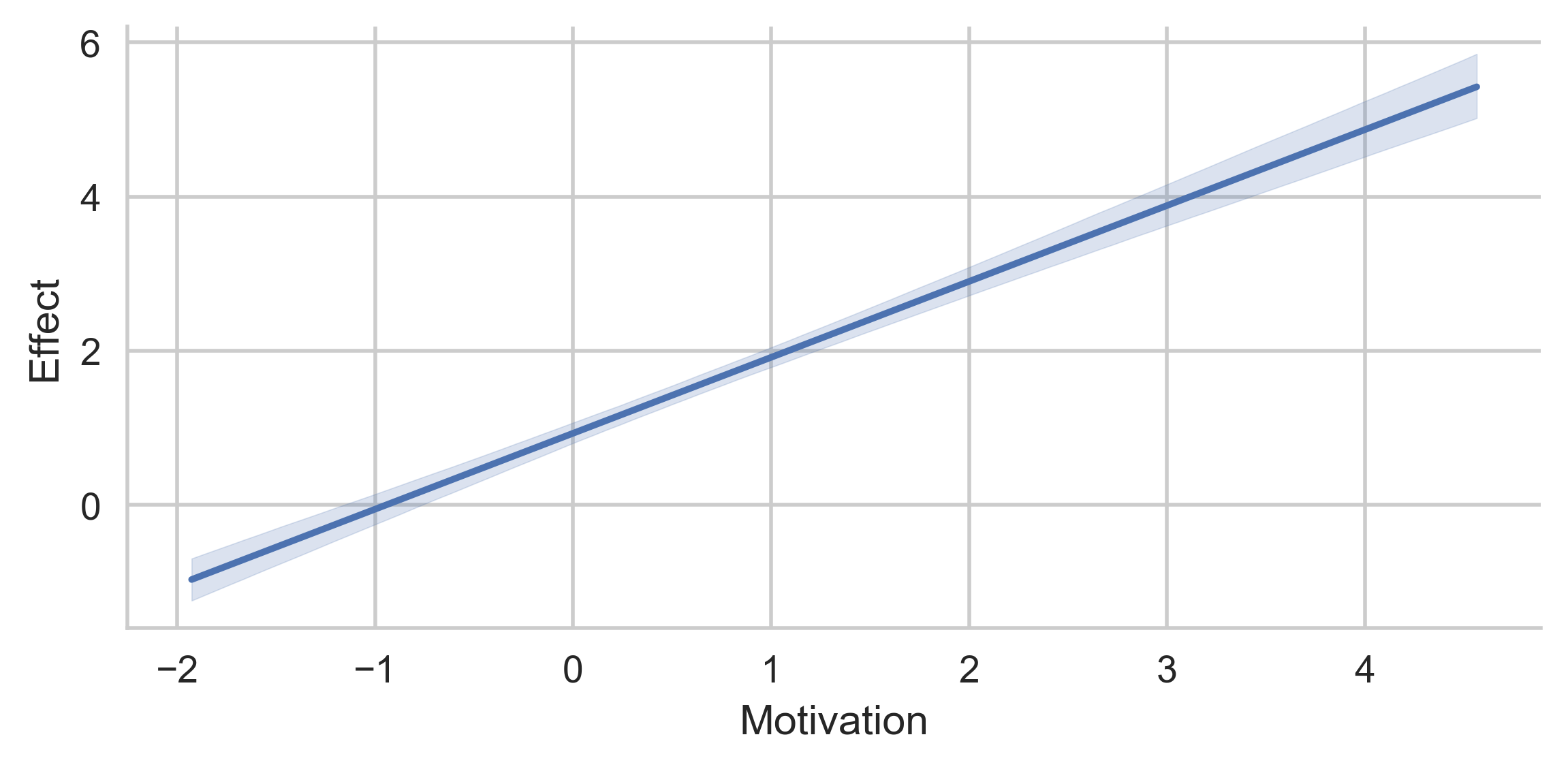
If you want the omitted moderator(s) to have a different value than 0,
you must pass a unique value for each moderator as a key in the
mods_at dictionary:
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation",
mods_at={"SkillRelevance":[-5]})
plt.show()
This plot represents the "partial" conditional indirect effect, when SkillRelevance is evaluated at -5.
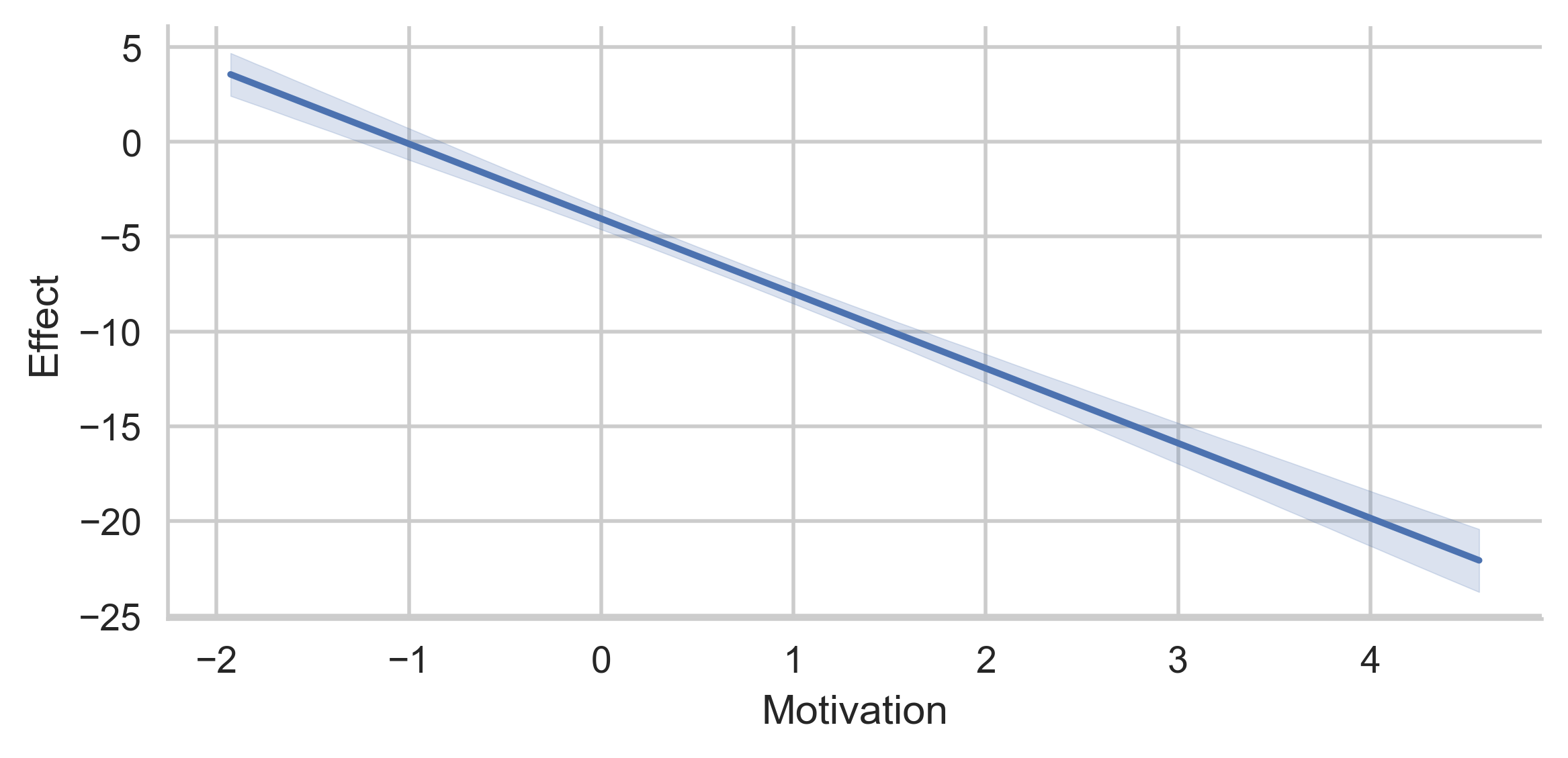
Do not pass multiple values in mods_at if you do not intend to
represent this moderator on the plot, as the graph will then not be
interpretable!
E. Customization
Under the hood, the plotting functions relies on a seaborn.FacetGrid
object, on which the following functions are called:
plt.plotwhenerrstyle="none"plt.plotandplt.fill_betweenwhenerrstyle="band"plt.plotandplt.errorbarwhenerrstyle="ci"
You can pass custom arguments to each of those objects to customize the appearance of the plot:
from pyprocessmacro import Process
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv("MyDataset.csv")
p = Process(data=df, model=13, x="Effort", y="Success", w="Motivation", z="SkillRelevance",
m=["MediationSkills", "ModerationSkills"], suppr_init=True)
# Plot: Make the lines bolder
plot_kws = {'lw': 5}
# Errors: Make the CI bolder and black
err_kws = {'capthick': 5, 'ecolor': 'black', 'elinewidth': 5, 'capsize': 5}
# Grid: Change the aspect ratio of the plot
facet_kws = {'aspect': 3}
g = p.plot_indirect_effects(med_name="MediationSkills", x="Motivation", errstyle="ci",
plot_kws=plot_kws, err_kws=err_kws, facet_kws=facet_kws)
plt.show()
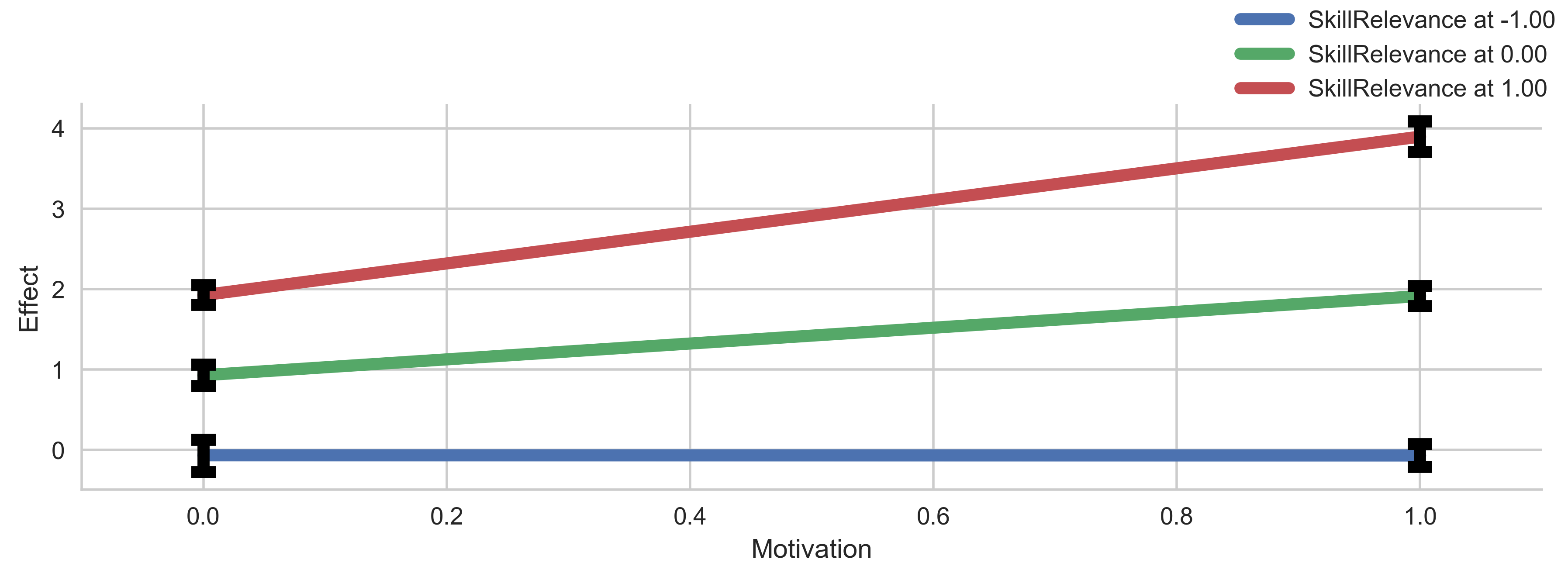
7. About
pyprocessmacro was developed by Quentin André during his Ph.D. in Marketing at INSEAD Business School, France.
His work on this library was made possible by Andrew F. Hayes' excellent book, by the financial support of INSEAD and by the ADLPartner Ph.D. award.
8. Copyright Notice
The original Process Macro for SAS and SPSS, and its associated files, are copyrighted by Andrew F. Hayes. The original code must not be edited or modified, and must not be distributed outside of http://www.processmacro.org.
Because pyprocessmacro is a complete reimplementation of the Process Macro, and was not based on the original code, permission was generously granted by Andrew F. Hayes to distribute pyprocessmacro under a MIT license.
This permission is not an endorsement of pyprocessmacro: all potential errors, bugs and inaccuracies are my own, and Andrew F. Hayes was not involved in writing, reviewing, or debugging the code.